New Progress in the Evolutionary Chloroplast Genomic Research of Ferns
2011-05-03
Important Progress in Evolutionary Chloroplast Genomic Research of Ferns
From Yuan Zhou et al., 2011, Population Genetics
The complex evolutionary history of the rpoB-psbZ region in ferns has been further clarified by the research group led by Professor Ting Wang from Wuhan Botanical Garden, Chinese Academy of Sciences with the cooperation of Sun Yat-sen University. This research has been published in BMC Plant Biology (2011, 11:64) entitled “Evolution of the rpoB-psbZ region in fern plastid genomes: notable structural rearrangements and highly variable intergenic spacers” (http://www.biomedcentral.com/1471-2229/11/64).
Lei Gao et al. from Wang’s group had published the first complete tree fern chloroplast genome of Alsophila spinulosa (Cyatheaceae) promoting our understanding of the evolutionary changes in fern chloroplast genomes (BMC Evolutionary Biology, 2009, 9:130). In that study, they observed that the rpoB-psbZ (BZ) region of some fern plastid genomes (plastomes) has experienced considerable genomic changes through fern evolution. However, the exact process of BZ evolution remains largely unknown.
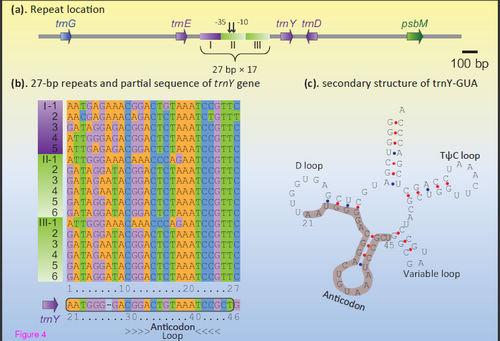
To address this issue, in the present study, Lei Gao et al. investigated a total of 24 fern BZ sequences with taxon sampling covering all the extant fern orders. They disclosed several new features of the evolutionary process of fern BZ regions, including (i) a tree fern Plagiogyria japonica contained a novel gene order that can be generated from either the ancestral Angiopteris type or the derived Adiantum type via a single inversion; (ii) the trnY-trnE intergenic spacer (IGS) of the filmy fern Vandenboschia radicans was expanded 3-fold due to the tandem 27-bp repeats which showed strong sequence similarity with the anticodon domain of trnY; (iii) the trnY-trnE IGSs of two horsetail ferns Equisetum ramosissimum and E. arvense underwent an unprecedented 5-kb long expansion, more than a quarter of which was consisted of a single type of direct repeats also relevant to the trnY anticodon domain; and (iv) ycf66 was independently lost at least four times in ferns.
This paper provided fresh insights into the evolutionary process of fern BZ regions. The most important aspect of this study is to provide evidence demonstrating that we need to consider the evolution of fern chloroplast genomes more complex than some previous studies implicated.