New Progress in the Molecular Evolution of Rubisco enzyme in Gymnosperms
2011-07-27
The complex molecular evolutionary patterns of the rbcL gene in gymnosperms have been illustrated by the research group led by Professor Ting Wang from Wuhan Botanical Garden, Chinese Academy of Sciences, with the cooperation of Dr. Mario A Fares of Dublin University and Prof. Yingjuan Su of Sun Yat-sen University. This research has been published in Biology Direct (2011, 6:29) entitled “Molecular evolution of rbcL in three gymnosperm families: identifying adaptive and coevolutionary patterns” (http://www.biology-direct.com/content/6/1/29).
The adaptive evolution of rbcL gene in three gymnosperm families, namely Podocarpaceae, Taxaceae and Cephalotaxaceae, is surveyed with the estimated timescale under the uncorrelated lognormal model together with the reconstructed phylogenetic relationships; meanwhile, conevolutionary analyses are also conducted using the software CAPS. The results indicate that: (1) Ten positively selected sites are detected in the gymnosperm RbcL subunit based on the random-site model and the branch-site model, implying that single copy chloroplast genes have a great feasibility of encountering adaptive evolution under ecological pressures. (2) The adaptive processes are identified at amino acid sites located on the contact regions among the Rubisco subunits and on the interface between Rubisco and its activase. The replacement of multiple amino acids at these regions may have optimized the holoenzyme activity during an episodic adaptation. This hypothesis rests on other piece of evidence from the coevolutionary analyses that support a correlated evolution between Rubisco and its activase, which seems to have accompanied the historic changing in the concentration of the atmospheric CO2. (3) A coevolutionary network is detected in the gymnosperm RbcL subunit. Moreover, six positively selected sites are found to be at the core nodes in the complex relationship. This finding shows that the large subunit of Rubisco can modify its structure and function through the coevolution of several amino acid sites and the modification will lead to the enzyme’s adaptive evolution.
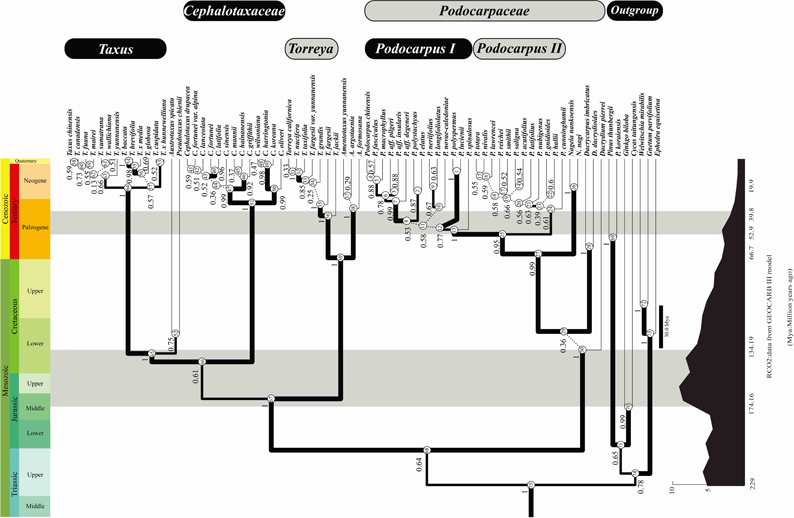
The phylogenetic tree of gymnosperm reconstructed relied on rbcL sequences(Image by Dr. Lin Sen)