Researchers Uncover the Molecular Mechanism of Grain Weight Regulation in Rapeseed
2015-09-08
WANG Hanzhong’s group from Oil Crops Research Institute of Chinese Academy of Agricultural Sciences, in collaboration with LI Rongjun from Wuhan Botanical Garden (CAS), reveals that the variation of auxin-response factor 18 (ARF18) genes on A9 chromosome in rapeseed can regulate the grain weight without changing the pod, which has an impact on production and laid the foundation for molecular design and cultivation of high yield varieties. It is the first polyploidy crop yield genes acquired by map-based cloning in the world and provides a reference for yield gene cloning in other major polyploid crops such as wheat, cotton etc. The world famous academic journal, the Proceedings of the National Academy of Sciences (PNAS), published the result online on August 31 entitled “Nature variation inARF18gene simultaneously affects seed weight and silique length in polyploidy rapeseed”.
70% value of major crops comes from seeds. The genetic improvement of seed production has been the focus of research and the functional gene cloning is also a hot spot. Seed yield is a quantitative trait controlled by multiple genes. In the important polyploid crops, such as wheat, rapeseed, cotton, etc., because of the complexity in genomes, map-based cloning of genes related to seed yield has been unable to get a breakthrough. In rapeseed, for example, although many teams in the world got a lot of major effect quantitative trait loci (QTLs) on grain weight in the past ten years, there is no report of the gene to be cloned.
With the significant difference in grain weight between rapeseed cultivars zy72360 and R1, this team constructed the segregating population and obtained a major effect QTL on the A9 chromosome, in which grain weight and pod length were collocated. The difference of grain weight in near isogenic lines (NILs) containing the main effect QTL was up to 20%. With the help of NILs and associated groups, the QTL interval was shortened to 50kb and 7 genes were found in this region. By genome sequence comparison between the parents, they found 4 genes which existed sequence variations. Transgenic analysis in Arabidopsis thaliana and Brassica napus ultimately identified that ARF18 was the target gene regulating seed weight and pod length. Overexpression of the gene could make the grain weight variation of 15%, while the seed number per pod kept no change.
Further studies showed that ARF18 was a transcription factor that regulated the expression of auxin responsive genes, and it has transcriptional inhibitory activity. By comparison of the ARF18 genes originated from the parental lines, they found the sequence variation of the 6th intron in the zy72360 induced abnormal shear which caused the loss of 7th exon, thus affected the function of the gene. Yeast hybrid and gel retardation experiments also demonstrated that the exon deletion in ARF18 in the large grain material resulted in the failure in dimer formation and the loss of inhibitory activity.
Seed is the offspring, and the pericarp is the mother's organ. In order to investigate the colocalization mechanism of grain weight, pod length and the relationship between each other, the NILs material was used for genetic research. Result showed that grain weight phenotype in the locus was regulated mainly by maternal genotype. Because of the pericarp photosynthesis provided the main carbon source in the late seed development, combined with gene expression patterns and transcriptome analysis, the study proposed that the loss of function in ARF18 in zy72360 led to longer pod, thereby increased the pericarp photosynthetic area. Longer pod provided more photosynthetic products for seed development which led to the increase of grain weight.
The study was funded by The National Key Basic Research Program of China (973), the National High Technology Research and Development Program of China (863), the National rapeseed industry technology innovation system and the innovation Engineering of Chinese Academy of Agricultural Sciences.
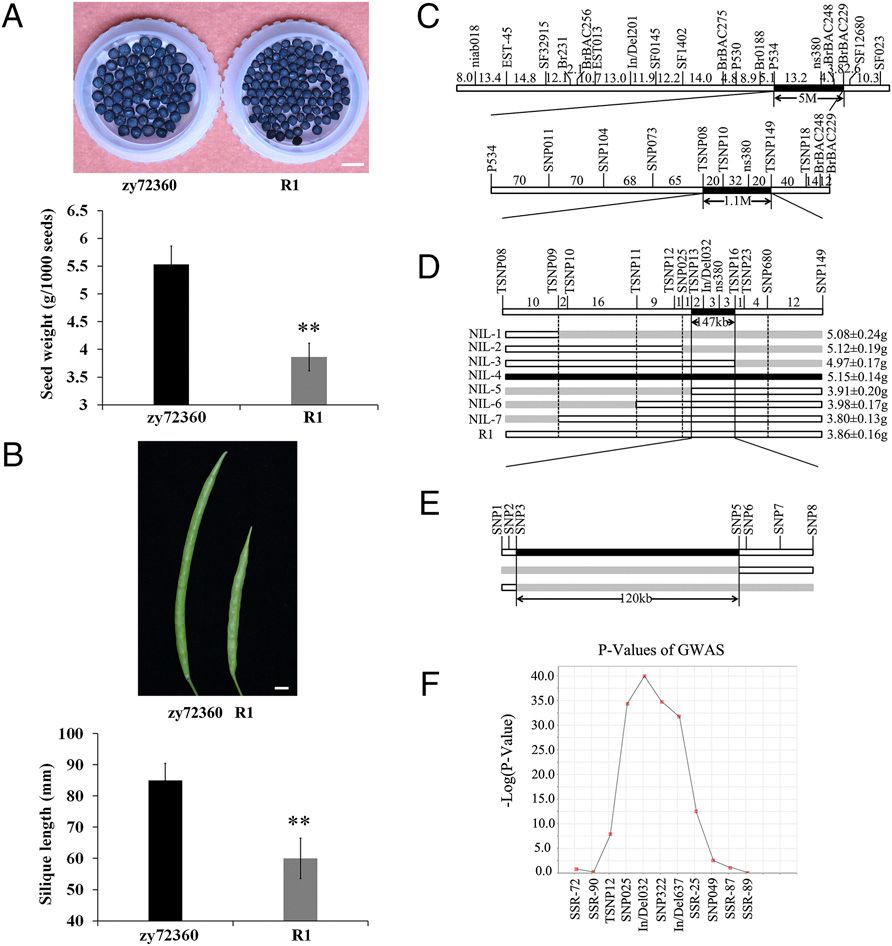
Fine mapping of the seed weight (SW) QTL in rapeseed using NILs and association populations (Image by WANG Hanzhong’s group)