Name:Yansheng Zhang
Tell:
Email:zhangys@wbgcas.cn
Organization:Wuhan Botanical Garden
De Novo Transcriptome Sequenced in P. lobata to Identify Putative Genes in Isoflavones Biosynthesis
2015-01-15
Pueraria lobata (Leguminosae) has long been used as a traditional Chinese herb. Several major isoflavones have been identified in the roots of P. lobata, such as daidzein, daidzin and puerarin, which have pharmacological effects. The presence of therapeutic isoflavones makes P. lobata an interesting plant in the recent study of isoflavones metabolism. However, the isoflavone metabolism and the characterization of corresponding genes in P. lobata remain largely unknown.
Given the extensive clinical usage and limited knowledge of the isoflavonoid biosynthesis in P. lobata, Prof. ZHANG Yansheng and his research team from Wuhan Botanical Garden constructed two cDNA libraries from root and leaf, and sequenced them by using Solexa (Illumina). Thus, about 13.3 G valid data were obtained and de novo assembled into 163,625 unigenes, among which 70,593 unigenes (43.1%) were aligned to the NCBI non-redundant (Nr) protein database.
RPKM (reads per kilo bases per million reads) method was used for comparing the differences of genes expression between roots and leaves. Compared to the root, 3148 unigenes were up-regulated, while 2011 genes were down-regulated in the leaf.
Gene ontology (GO) and pathway enrichment analyses were also performed to further understand the differentially expressed genes. Results revealed that 47 novel unigenes were found in the biosynthesis of isoflavones.
22 putative UDP glycosyltransferases (UGTs) and 45 O-methyltransferases (OMTs) unigenes expressed at a high level in the roots compared to leaves, likely involved in the tailoring processes of isoflavonoid downstream pathway.
Analysis of MYB transcription factors (TFs) showed that 133 MYB encoding genes were upregulated in the root, and they were probably candidate regulators for isoflavonoid biosynthesis in P. lobata.
In this research, the de novo transcriptome investigation of these unique transcripts facilitates functional studies of isoflavones biosynthesis in P. lobata and will improve the medicinal ingredients production through metabolic engineering in the future.
Results entitiled “De novo transcriptome sequencing inPueraria lobatato identify putative genes involved in isoflavones biosynthesis” were published in Plant Cell Report.
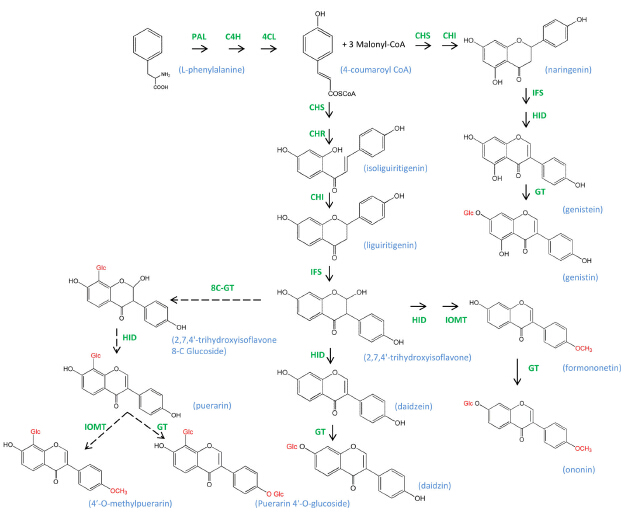
Predicted isoflavonoid biosynthetic pathway-related genes and products in Pueraria lobata (Image by Dr. WANG Xin)