Name:HU Guangwan
Tell:
Email:guangwanhu@wbgcas.cn
Organization:Wuhan Botanical Garden
Researchers Analyze the Complete Chloroplast Genomes of Rhipsalis baccifera, the Only Cactus with Natural Distribution in the Old World
2020-10-20
Family Cactaceae is almost endemic to the New World except for the epiphytic Rhipsalis baccifera (J.S.Muell.) Stearn, which is found naturally occurring in both the New and the Old Worlds. This species has thus drawn the attention of most researchers. However, there is still insufficient molecular work on R. baccifera and also so far, there is no work on the complete chloroplast genome analysis of this species.
Flora and Plant Taxonomy in Eastern Africa Research Group of CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture at Wuhan Botanical Garden analyzed for the first time the complete chloroplast genomes R. baccifera collected from Kasigau forest, Kenya.
The complete chloroplast genome of R. baccifera has a total length of 122,333 base pairs (bp) in size. It displays a typical quadripartite structure with a large single-copy (LSC) region of 81,459 bp in length, separated from the SSC region of 23,531 bp, by two inverted repeat (IR) regions both 8530 bp long. The genome contains 110 genes, with 73 protein-coding genes, 31 tRNAs, four rRNAs and two pseudogenes (Figure 1).
Comparative analysis with eight species of the ACPT (Anacampserotaceae, Cactaceae, Portulacaceae, and Talinaceae) clade of the suborder Portulacineae species reveals an unique inversion and rearrangement only observed in R. baccifera at the LSC region for the first time, representing the most significant structural change in terms of its size (Figure 2). Inversion of the SSC region seems common in subfamily Cactoideae, and another 6 kb gene inversion between rbcL- trnM is observed in R. baccifera and Carnegiea gigantea.
The phylogenetic analysis involving 36 species of the order of Caryophyllales and two outgroups supports monophyly of the families of the ACPT clade. R. baccifera occupies a basal position in the clade formed by Cactaceae species. A high number of rearrangements in this cp genome suggest a larger number of mutation events in the history of evolution of R. baccifera. (Figure 3)
These results provide important tools for future work on R. baccifera and in the evolutionary studies of the suborder Portulacineae.
The research entitled “Complete Chloroplast Genome of Rhipsalis baccifera, the only Cactus with Natural Distribution in the Old World: Genome Rearrangement, Intron Gain and Loss, and Implications for Phylogenetic Studies” was published in Plants. This work was supported by the National Natural Science Foundation of China and Sino-Africa Joint Research Center, CAS.
Kenyan student Millicent Akinyi Oulo, together with two Chinese students: YANG Jiaxin and DONG Xiang are the co-first authors. Professor HU Guangwan is the corresponding author. Several students from the group participated in the study.
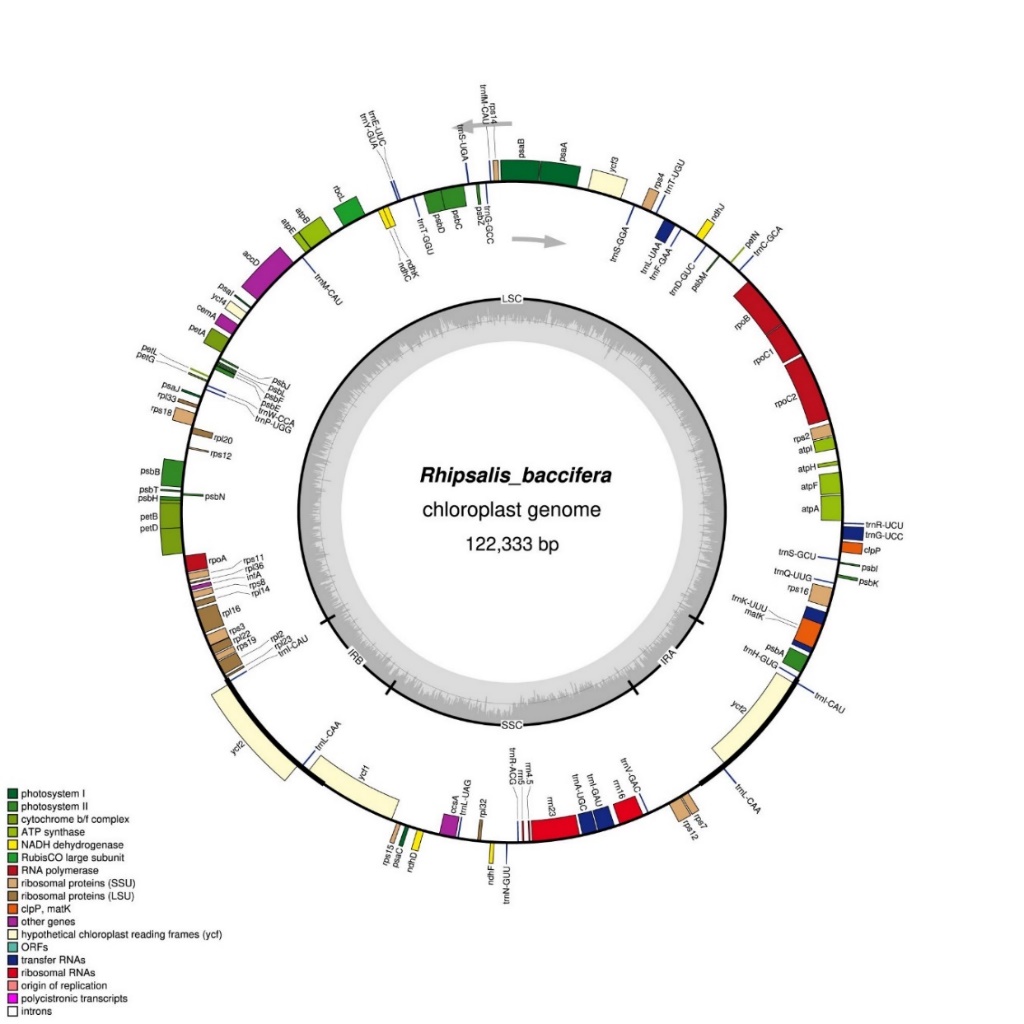
Figure 1. Complete chloroplast genome of Rhipsalis baccifera (Image by WBG)

Figure 2. A 19-gene inversion in the LSC region of R. baccifera between genes ndhJ- trnY. The cp genome of R. baccifera relative to Spinacia oleraceae (Image by WBG)

Figure 3. Combined phylogenetic tree of 36 species order Caryophyllales using maximum likelihood (ML) and Bayesian methods based on their complete chloroplast genomes (Image by WBG)