Name:LI Li
Tell:
Email:lili@wbgcas.cn
Organization:Wuhan Botanical Garden
Rhizobium Adapts Lifestyles from Rhizosphere to Symbiosis
2020-12-03
Rhizobia are soil-dwelling bacteria that form symbioses with legumes and provide biologically useable nitrogen as ammonium for the host plant. During symbiosis, rhizobia must adapt to several different lifestyles. These range from free-living growth in the rhizosphere, through root attachment and colonization, to passage along infection threads, differentiation into bacteroids that fix N2, and, finally, bacterial release from nodules at senescence.
Researchers presented an in-depth investigation of N2 - fixing bacterial interactions with a host legume during symbiosis by examining bacterial gene requirement at different stages of rhizosphere growth, root colonization, and nodulation by using multistage mariner transposon - insertion sequencing (INSeq).
A total of 603 genetic regions were required to competitively nodulate peas. Of these, 146 were classified as rhizosphere - progressive, required not only in the rhizosphere but also at every subsequent stage, highlighting how critical successful competition in the rhizosphere was to subsequent infection and nodulation. As expected, 211 genes were required by both nodule bacteria and bacteroid function.
There were 17 genes required only in the rhizosphere, and 23 genes specifically required for root colonization.
These results dramatically highlight the importance of competition at multiple stages of a Rhizobium–legume symbiosis.
Results were published on PNAS entitled “Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis”. University of Oxford and Wuhan Botanical Garden were the co-research units. Associate Prof. LI Li from Wuhan Botanical Garden is the co-first author. The research was supported by the fund project of China Scholarship Council.
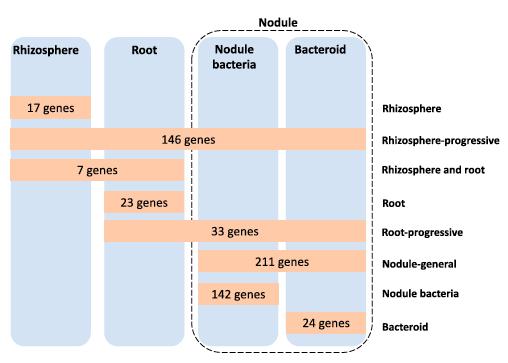
Rlv3841 genes required during the stages of symbiosis with pea (Image by Oxford and WBG)
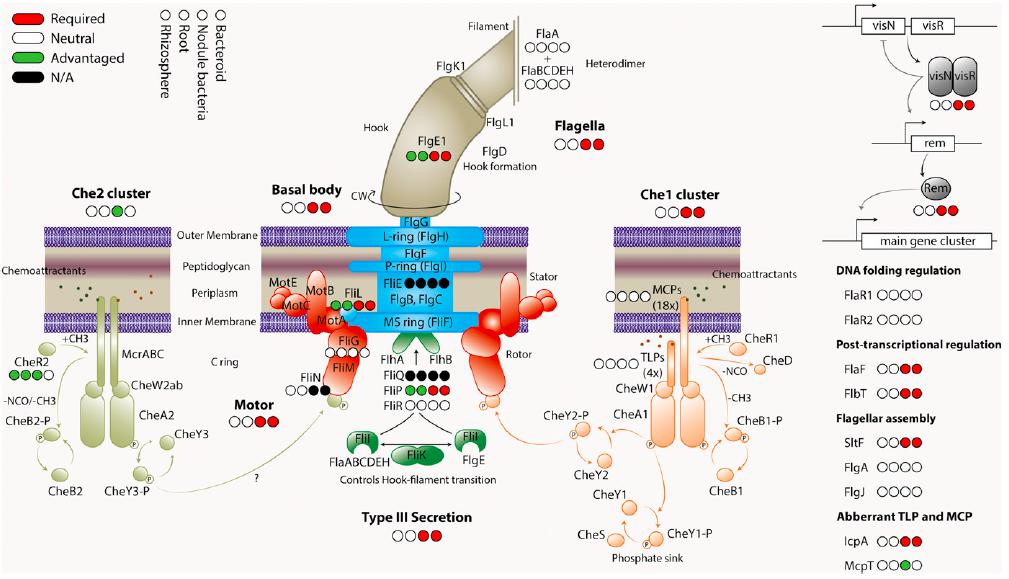
Classification of genes encoding motility and chemotaxis proteins in rhizosphere, root, nodule bacteria, and bacteroid INSeq libraries (Image by Oxford and WBG)
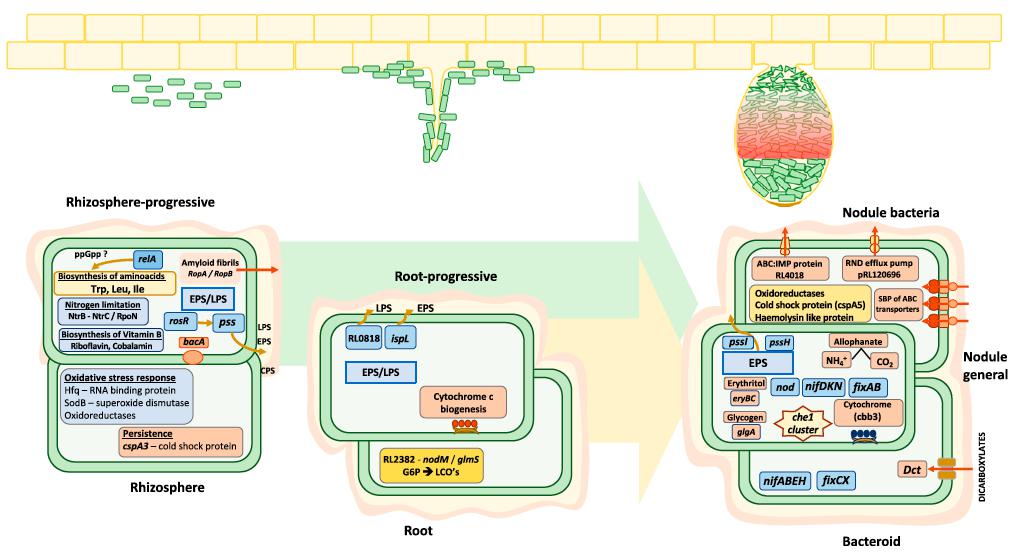
Rlv3841 genes required at different stages of symbiosis with pea from INSeq analysis (Image by Oxford and WBG)