Name:Lei Gao
Tell:
Email:leigao@wbgcas.cn
Organization:Wuhan Botanical Garden
Genome of Solanum pimpinellifolium Brings Wild Genomic Reservoir for Tomato Breeders
2020-12-22
The progenitor of cultivated tomato, Solanum pimpinellifolium (SP), is an important germplasm donor in modern tomato breeding and a widely used model species for tomato genetic and developmental research. Compared to the cultivated tomato, SP displays many desirable traits such as higher tolerance to biotic and abiotic stress, more intense flavor and higher lycopene levels. SP genome assembly and annotation are needed for studying the genetics and molecular mechanisms underlying these traits.
The research team, led by Prof. FEI Zhangjun from Boyce Thompson Institute, with assistance from Prof. GAO Lei from Wuhan Botanical Garden, constructed a high-quality chromosome-scale reference genome assembly of SP accession LA2093 using PacBio long reads combined with Hi-C chromatin interaction maps.
Structural variants (SVs) are known to underlie many domestication-related phenotypes. Although whole genome SNPs have been used to reconstruct the history of tomato domestication and to study the impact of human selection on the tomato genome, the population dynamics of SVs in tomato is largely unexplored.
In this study, more than 92,000 high-confidence SVs were identified between the genomes of SP LA2093 and cultivar Heinz 1706. The SVs were further genotyped in ~600 tomato accessions, representing SP, S. lycopersicum var. cerasiforme (SLC), and S. lycopersicum var. lycopersicum (SLL) heirloom and modern varieties. Numerous SVs underlying important breeding traits, such as fruit weight, lycopene metabolism, ripening, sugar metabolism and disease resistance, were discovered to be under selection during tomato domestication and breeding.
Expression quantitative trait locus (eQTL) analysis was further employed to investigate the regulatory roles of SVs using tomato fruit transcriptome data. A total of 48 distant-acting eQTL hotspots, as well as the potential master regulators within them, were identified. The results confirmed the previous finding that MYB12 is the key regulator of flavonoid biosynthesis in fruit and identified novel SVs contributing to this complex regulatory network.
In addition, an AP2/ERF transcription factor orthologous to Arabidopsis WRINKLED 3 (WRI3), in one major distant-acting eQTL hotspot, was identified as the master regulator targeting multiple lipid biosynthetic genes expressed in the epidermis of developing fruit. This result provides knowledge of the regulatory mechanism involved in fruit cuticular lipid accumulation.
The LA2093 genome sequence and the SVs identification provides rich resources for future research and breeding programs to improve fruit quality and stress tolerance.
The results have been published in Nature Communications entitled “Genome ofSolanum pimpinellifolium provides insights into structural variants during tomato breeding”.
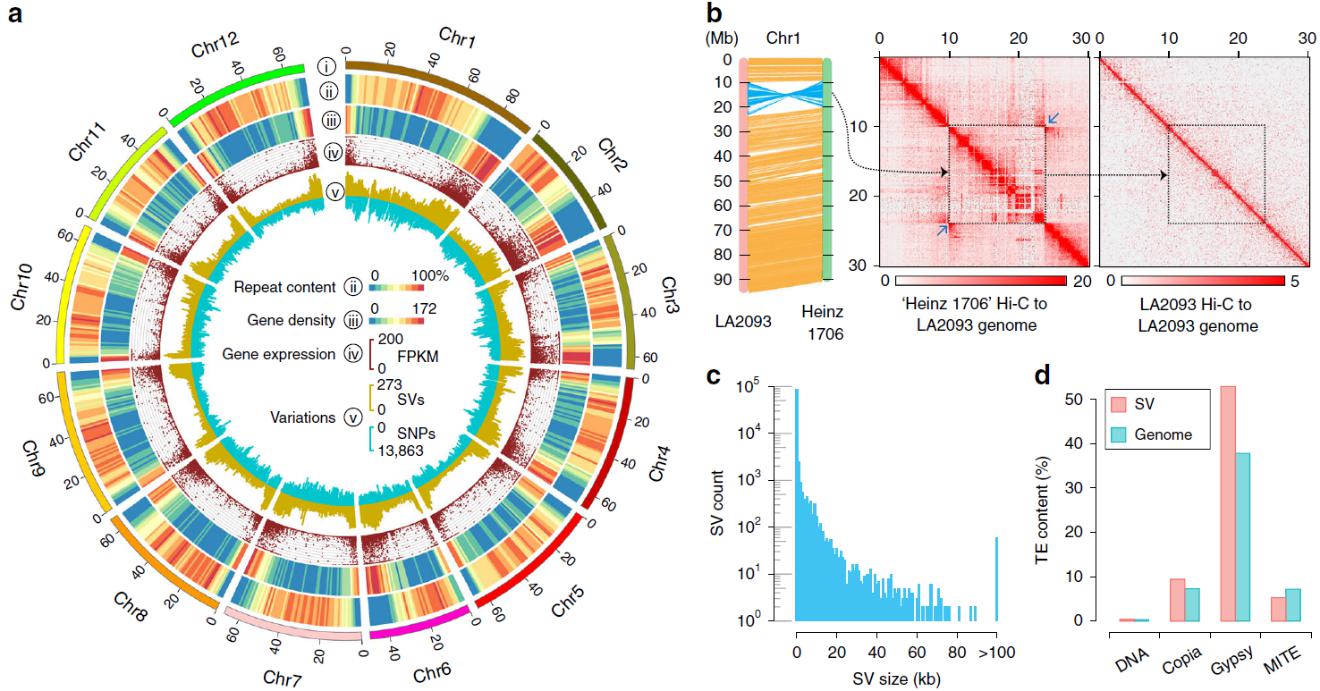
Genomic landscape of S. pimpinellifolium LA2093 and structural variants identified between LA2093 and Heinz 1706 (Image by Boyce Thompson Institute and WBG)