Name:Tao Shi Jinming Chen
Tell:
Email:shitao323@wbgcas.cn jmchen@wbgcas.cn
Organization:Wuhan Botanical Garden
Researchers Reappraise the Genome Duplication in the Ancestral Eudicot
2021-01-19
Eudicot is one of the major branches of the flowering plants. Basal eudicots (early-diverging) include Ranunculales, Proteales, Trochodendrales and Buxales. Recent chromosomal level genome assemblies of Aquilegia, Papaver, sacred lotus and Tetracentron can provide important clues for genome evolution of eudicot ancestor.
Early studies on grape and cacao suggest that the ancestral core eudicot experiences a genome triplication. Recently, Aköz and Nordborg found that the basal eudicot, Aquilegia was a tetraploid. Based on similarity clustering of gene order from Aquilegia, grape and cacao, they suggested that the ancient tetraploidy was shared between Aquilegia and core eudicots. They further suggested that the genome hexaploidy γ was formed by hybridization and followed by a genome duplication (Aköz & Nordborg, 2019, Genome Biology, 20:256)
Researchers from Wuhan Botanical Garden, SHI Tao and CHEN Jinming found that Aköz and Nordborg’s conclusion seemed to be problematic. This is because, their previous study on another basal eudicot, lotus, did not identify Aköz Nordborg’s hypothesized the ‘eudicot-wide tetraploidy’, but instead, a lotus-specific tetraploidy occurring after its divergence to Macadamia (another Proteales plant) (see Shi et al. 2020, Molecular Biology and Evolution, 37(8): 2394-2413).
SHI and CHEN found a rapid decay of Aquilegia intra-specific synteny, which showed more loss than earlier inter-specific syntenies, Aquilegia-Nympheae and Aquilegia-Ceratophyllum. Therefore, Aköz & Nordborg’s simple clustering of gene order did not consider the rate heterogeneity of the synteny loss rates between intra- and inter-species.
The similarity of genome structure between grape and Aquilegia is likely as a consequence of homoplasy of genome evolution after polyploidy, such as dosage balance and selection of single-copy genes.
Meanwhile, SHI and CHEN also found Aköz & Nordborg’s another conclusion that a common chromosomal fusion in the tetraploid eudicot ancestor was unlikely, as SHI and CHEN found the fused chromosomes in grape and Aquilegia did not share the most structural similarity but shared a higher similarity of the other non-fused homologous regions. SHI and CHEN finally performed phylogenomic analyses and validated that the Aquilegia genome duplication was only lineage-specific rather than eudicot-wide.
To sum up, this study on the genome duplication of eudicot sheds light on the genome evolution of early eudicots.
This article entitled “A reappraisal of the phylogenetic placement of the Aquilegia whole-genome duplication” is now published in Genome Biology.
This project was supported by grants from the Strategic Priority Research Program of Chinese Academy of Sciences, the National Natural Science Foundation of China, and Youth Innovation Promotion Association of Chinese Academy of Sciences.
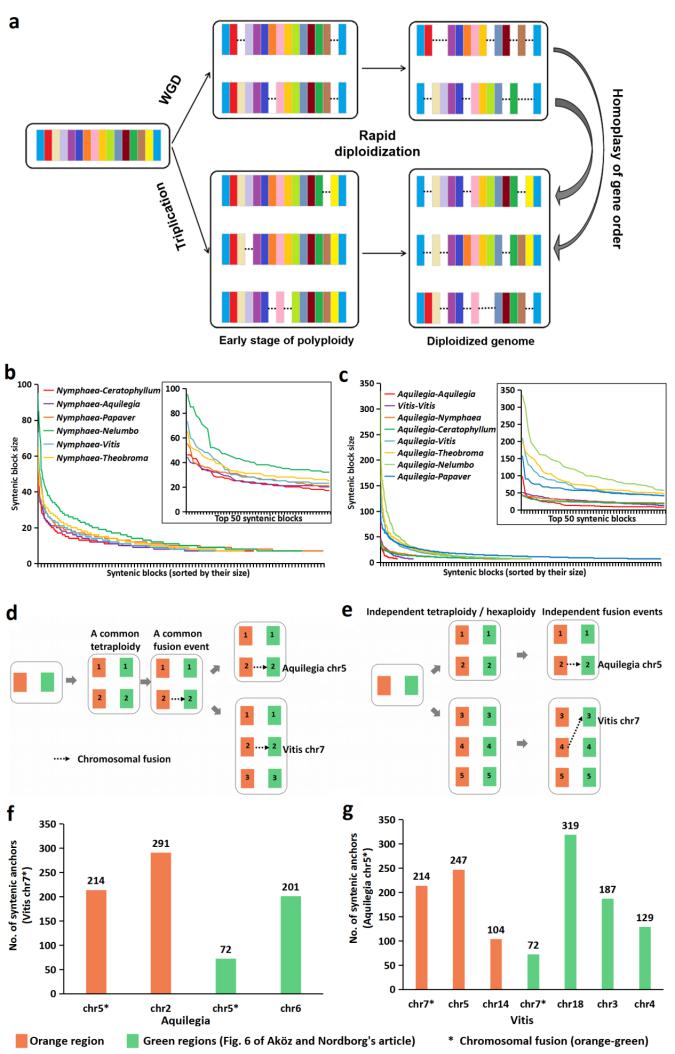
Evidence of extensive loss of Aquilegia intra-specific synteny and prediction of two independent chromosomal fusion events (Image by WBG)