Name:HU Guangwan
Tell:
Email:guangwanhu@wbgcas.cn
Organization:Wuhan Botanical Garden
Complete Chloroplast Genome Sequence of Fortunella venosa Provides Robust Support for Its Status as An Independent Species
2021-10-12
Fortunella venosa (Champ. ex Benth.) C.C.Huang is a perennial evergreen shrub in the family Rutaceae. This species is endemic to China, having an overlapping distribution with the tetraploid species F. hindsii (Champ. ex Hook.) Swingle (FOC: Citrus japonica Thunb.), with distribution range occupying Nanping (Fujian), Yongfeng (Jiangxi), and recently found in Chaling, Ningyuan, Guidong, and other Counties in Hunan Province. Its wild population has been under threat due to climate change and anthropogenic pressures, thus it was listed as an endangered species in China Species Red List (Vol.1 Red List). Besides, its placement has been controversial basing on its taxonomy and systematics. In Flora of China (FOC), the genus Fortunella was incorporated into the genus Citrus, with some of its species being treated as synonyms of C. japonica Thunb.
Researchers from the College of Life Sciences of Hunan Normal University, together with the Flora and Plant Taxonomy in Eastern Africa Research Group of Wuhan Botanical Garden, CAS, sequenced and analyzed the complete chloroplast (cp) genome of Fortunella venosa collected from Zhuzhou region, Hunan Province, China, and did both molecular and morphological comparative analysis with Citrus japonica Thunb.
The Chloroplast genome size of Fortunella venosa is 160,265 bp, with a typical angiosperm four-part ring structure consisting of a large single copy region (87,597 bp), a small single copy region (18,732 bp), and two inverted repeat regions (26,968 bp each). There are 134 predicted genes in Chloroplast genome, including 89 protein-coding genes, 8 rRNAs, and 37 tRNAs (Figure 1).
The phylogenetic results of this study (Figures 2 and 3) indicate that Fortunella Swingle and Citrus L. are closely related, but do not support the incorporation of F. venosa into C. japonica. It shows that they are two different species basing on the phylogenetic analysis.
This study shows that Fortunella venosa should be an independent species, which is significantly different from F. japonica in terms of morphology (habitat, leaf type, fruit size, etc.). F. venosa is a small shrub, usually no more than 1 m tall (the shortest mature plant is 0.28 m high); the leaves are single leaves (the joints of the petiole and the leaf are not joint); the leaves are usually small, 2-4 (- 7) cm long, 1-2 cm wide, wedge-shaped base; petiole short, 1-3 (- 5) mm long; flower solitary leaf axils, petals 3- 5 mm long; ovary 2- 3 compartments; fruit spherical or elliptical, diameter 6- 8 mm, Orange- red when mature. On the other hand, F. japonica is a small tree or tree with a height of 2 to 8.5 m, and the main stem is usually slender; the leaf is a single leaflet with joints between the petiole and the leaf; the leaf is larger, 4- 9 cm long, 1.5- 3.5 cm wide, and a wide wedge-shaped base; The petiole is obviously longer, 6- 10 (- 15) mm long; the flower is single or 2- 3 clusters with leaf axils, the petals are 6- 8 mm long; the ovary is 4- 6 compartments; the fruit is larger, spherical, 1.5- 2.5 cm in diameter, orange- yellow to orange- red when cooked (Figure 4).
The research entitled “Complete Chloroplast Genome Sequence ofFortunella venosa (Champ. ex Benth.) C.C.Huang (Rutaceae): Comparative Analysis, Phylogenetic Relationships, and Robust Support for Its Status as an Independent Species” was published in the international journal of Forests, section Genetics and Molecular Biology.
This work was supported by grants from the National Science and Technology Basic Research Survey Project, Investigation of Forest Tree Germplasm Resources in Hunan Province, and International Program of Chinese Academy of Sciences. WANG Ting was the first author, and Prof. HU Guangwan and Dr. CAI Xiuzhen were the corresponding authors.
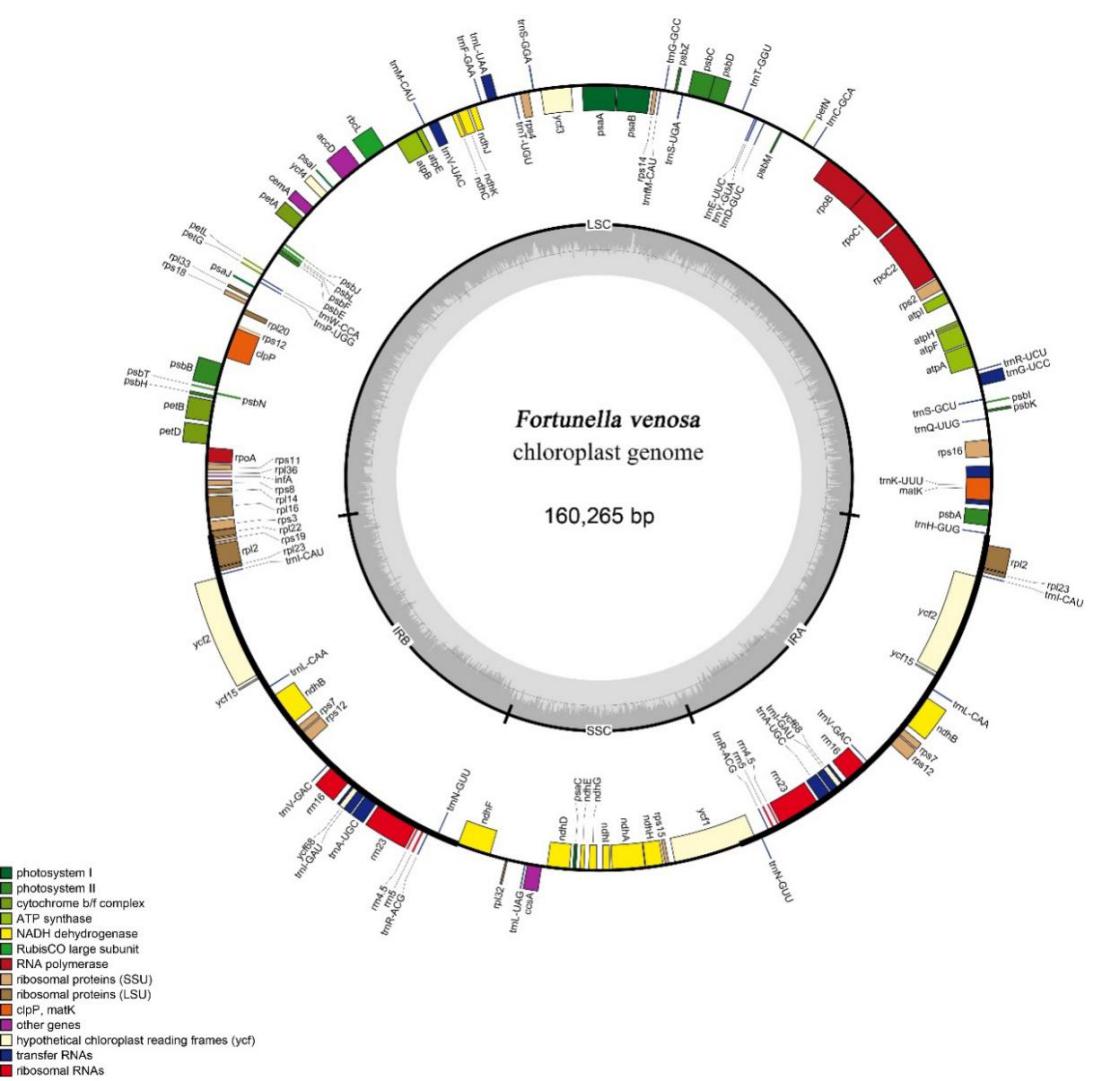
Figure 1. Gene map of the Fortunella venosa chloroplast genome. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise. Genes belonging to different functional groups are color coded. The darker gray color in the inner circle corresponds to the GC content, and the lighter gray color corresponds to the AT content (Image by WANG Ting )
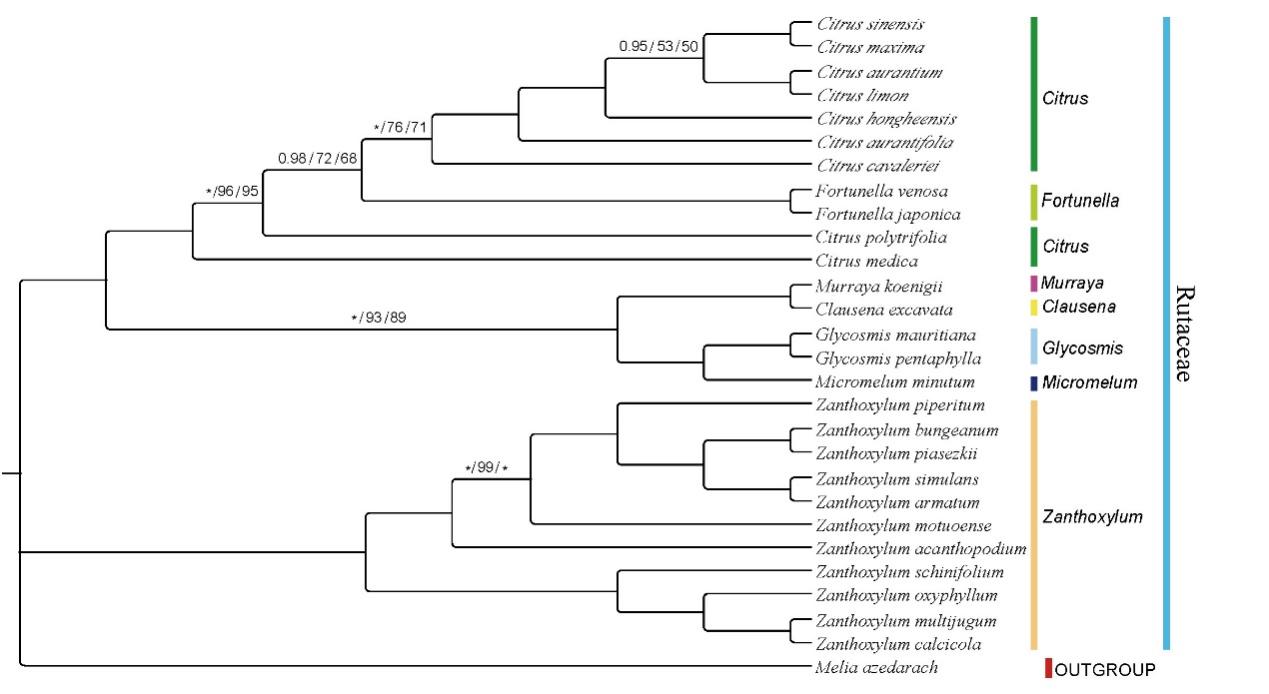
Figure 2. Phylogenetic tree generation using 76 CDS common in 27 Rutaceae species. Melia azedarach was used as outgroups. The numbers above the branch represent bootstrap support value for BI/ML/PhyML methods, where the asterisk signifies maximum support value of 100 in IQ and 1 BI. Blank branches signify 100% support value (Image by WANG Ting )
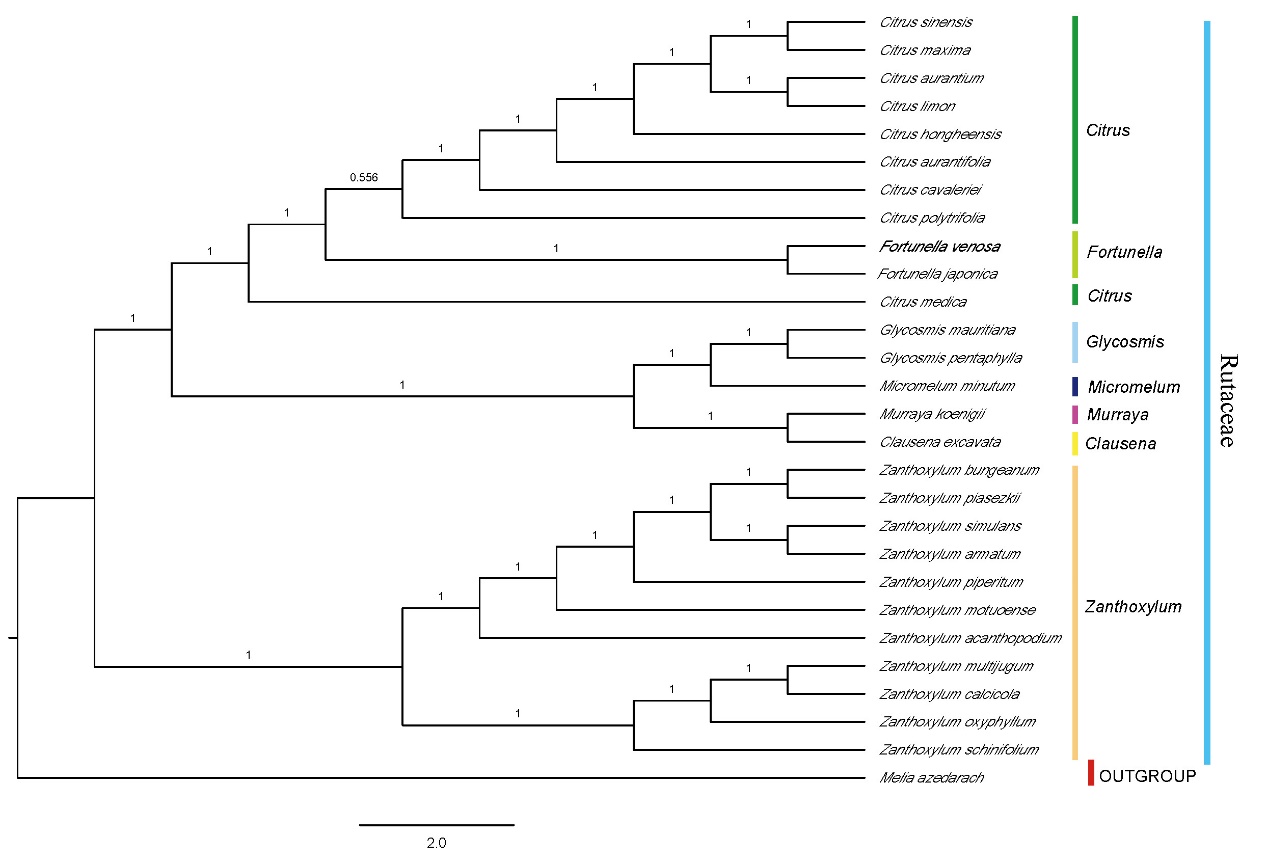
Figure 3. Phylogenetic trees based on BI of Rutaceae species based on whole chloroplast genome sequences, with one species from family Meliaceae used as outgroup. The Bayesian inference (BI) tree with posterior probabilities values on the branches (Image by WANG Ting )

Figure 4. Main morphological forms of F. japonica and F. venosa. A. F. japonica; B. F. venosa. 1. Plants and habitats; 2. Branches; 3. Leaves; 4. Flowers; 5. Fruit branches; 6. Fruits and fruits cross-cut (Image by WANG Ting )