Name:HU Guangwan
Tell:
Email:guangwanhu@wbgcas.cn
Organization:Wuhan Botanical Garden
Research Reveals the Plastome Evolution and Phylogenetic Relationships of Apocynaceae
2023-03-01
Apocynaceae, the dogbane family, is one of the 10 largest families in angiosperms, comprising 378 genera and more than 5000 species. However, the backbone phylogeny of the family is less well supported, and the evolutionary pattern of plastomes in this large family is much unclear.
To have a better understanding of the phylogenetic relationships and plastome evolutionary pattern of Apocynaceae, researchers of Group of African Plant Taxonomy and Flora from Wuhan Botanical Garden obtained a total of 101 complete plastomes, representing 26 of 27 tribes of Apocynaceae, to conduct comparative plastome analyses. Phylogenetic trees were also reconstructed using maximum likelihood and Bayesian inference methods.
As a result, the 101 Apocynaceae complete plastomes showed a typical quadripartite structure, including a large single copy (LSC) region, a small single copy (SSC) region, and two inverted repeated (IR) regions. Plastome lengths ranged from 150,897 bp in Apocynum venetum L. to 178,616 bp in Hoya exilis Schltr., with a difference of 27,719 bp. Six types of IR/SC boundaries were found in the 101 Apocynaceae plastomes. Different sizes of IR expansion were found in three lineages (Alyxieae, Ceropegieae and Marsdenieae), suggesting that IR expansions have occurred independently at least three times in Apocynaceae. Two clades within Marsdenieae showed different trends in IR expansion direction: the “Cosmopolitan Clade” expands towards LSC, while the “Asia-Pacific Clade” towards both LSC and SSC.
Five non-coding regions and six protein-coding genes were found to be highly diverse: accD, ccsA-ndhD, clpP, matK, ndhF, ndhG-ndhI, trnG(GCC)-trnfM(CAU), trnH(GUG)-psbA, trnY(GUA)-trnE(UUC), ycf1, and ycf2. They can be used as promising candidate markers for shallower phylogenetic analyses and DNA barcoding.
Maximum likelihood and Bayesian phylogenetic analyses yielded nearly identical tree topologies. A total of 15 consecutive dichotomies were identified along the backbone. The subfamily Periplocoideae were nested in the Apocynoid grade and were sister to a clade of Echiteae-Odontadenieae-Mesechiteae clade with strong support.
The placement of Periplocoideae, and non-monophyletic Willughbeieae, Melodineae, Hoya, and Ceropegia, as well as those conflict relationships on nuclear phylogeny, suggest there is much room for improvement on the phylogeny and taxonomy of Apocynaceae at ranks above genus.
This study assembled the largest plastome dataset to date for Apocynaceae, studied the plastome evolution for the first time across the whole family, and reconstructed a phylogeny with a well-resolved backbone.
The research was supported by the Biological Resources Program, Chinese Academy of Sciences(CAS), China National Plant Specimen Resource Center, International Partnership Program of CAS, and Sino-Africa Joint Research Center, CAS.
The results have been published in Molecular Phylogenetics and Evolution entitled "Evolution of 101 Apocynaceae plastomes and phylogenetic implications".
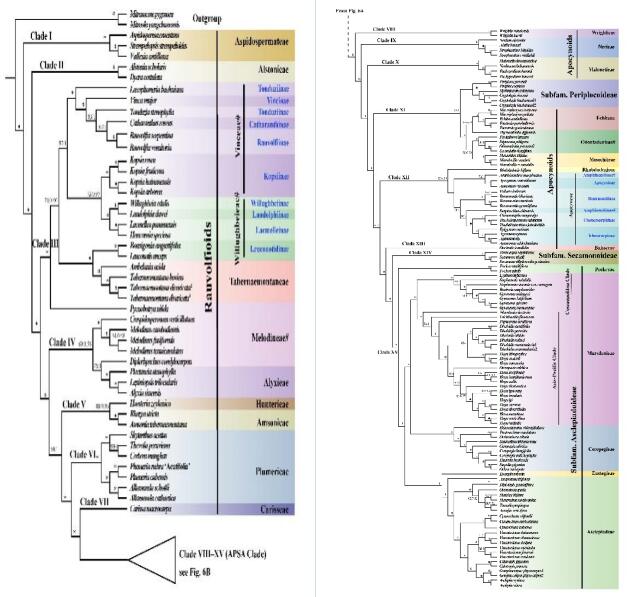
Consensus phylogenetic tree reconstructed by maximum likelihood (ML) and Bayesian inference (BI) analysis based on 77 protein-coding sequences (CDS) of 162 Apocynaceae species and two outgroups (Image by WANG Yan)
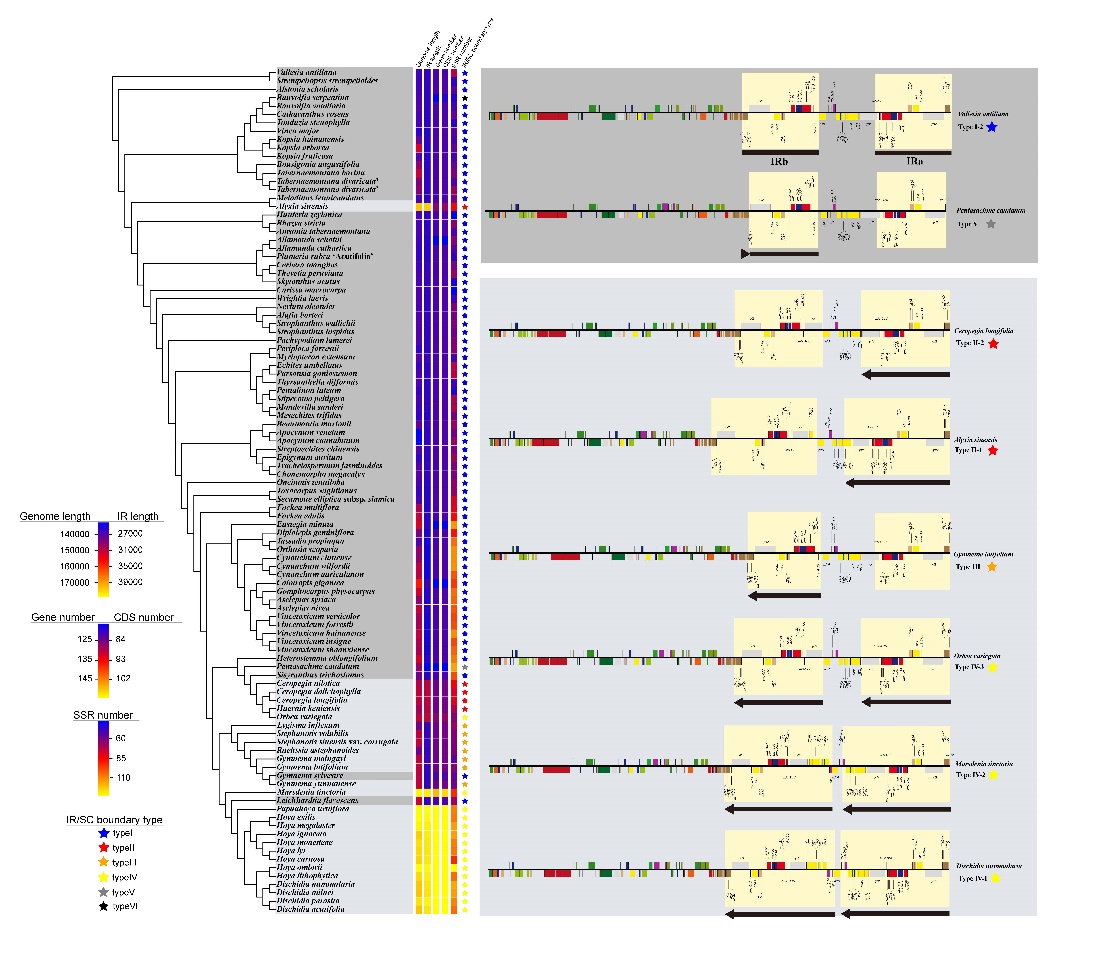
Plastome characters mapped to topology (Image by WANG Yan)