Name:ZHANG Xiujun
Tell:
Email:zhangxj@wbgcas.cn
Organization:Wuhan Botanical Garden
Study Unveils the Puzzling Nature of RNA Editing in Plants
2023-05-09
RNA (Ribonucleic Acid) editing is an important process to maintain essential functions of encoded proteins at the RNA level. Recent studies reveal that RNA editing is a widespread phenomenon that occurs in various land plants, including the liverworts, mosses, hornworts, lycopods, ferns, and flowering plants. However, few comparison study has been conducted in terms of intra- and inter-groups for chloroplast RNA editing except for ferns.
To gain a better understanding of RNA editing in plant chloroplast, researchers from Wuhan Botanical Garden of the Chinese Academy of Sciences chosed diverse plant species that represented three distant evolutionary clades (fern, gymnosperm, and angiosperm), and determined thousands of editing sites based on the amount of RNA-seq data.
Results reveal a total of 5203 editing sites located in chloroplast genes across 21 species after rigorous screening. The clustering relations of numbers of RNA editing sites agree with the phylogenetic tree based on gene sequences approximately, verifying that RNA editing across the plant kingdom is comparatively conservative and accords with laws of evolution roughly.
The numbers of editing sites decline with the evolution of plants, and editing events occur more often in the early diverging plant than later-branching ones for each clade, which implies that RNA editing may break out in early branching plants from different clades simultaneously and suffer a lot of loss during evolution.
Additionally, the distribution of editing levels among three aspects (condon positions, species and edited genes) is also explored. It is found that the average RNA editing level varies widely across 21 species as well as genes, with the lowest as 0.42 in Selaginella moellendorffii, and the highest up to 0.88 in the atpA gene (ATP synthase alpha-subunit gene). The declination of cytosine content with evolution detected in this study further indicates that situation of genomic sequence is the significant driver of loss of editing for later-branching plants.
These results reveal the diversity of RNA editing in chloroplast transcripts across three main plant clades, many of the identified sites in the study have not been previously reported, and it will help understanding the puzzling nature of RNA editing in plants.
This study has been published in Plant Systematics and Evolution entitled "Diversity of RNA editing in chloroplast transcripts across three main plant clades". (Published online: 28 March 2023)
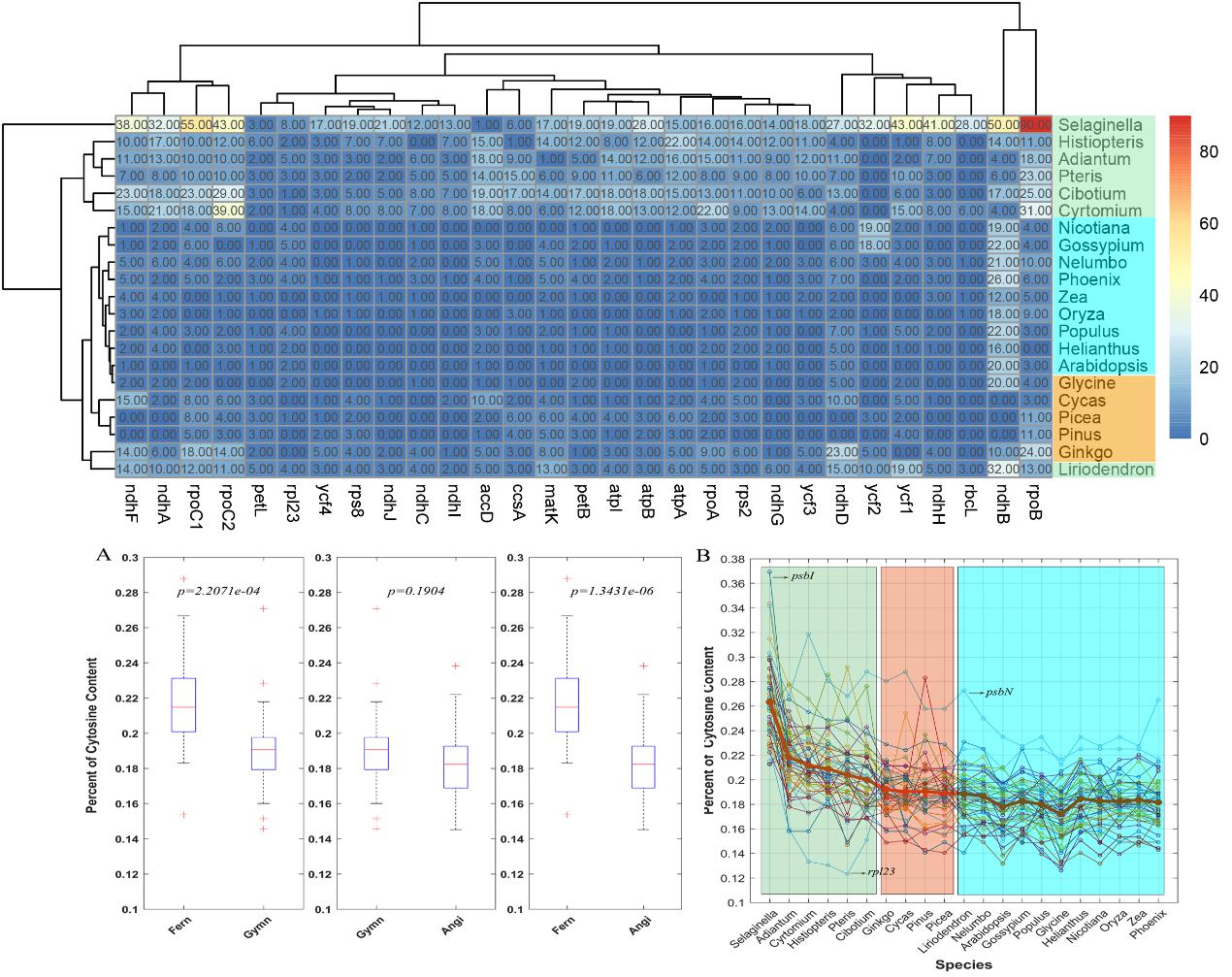
The hierarchical cluster of numbers of RNA editing sites and statistics of cytosine content in shared edited genes across 21 species (Image by WBG)